Professor

| Email: | lindahl@umbc.edu |
|---|
Education
Postdoctoral, Molecular Biology, University of Wisconsin, 1976
Ph.D., Microbiology, University of Copenhagen, Denmark, 1973
Professional Interests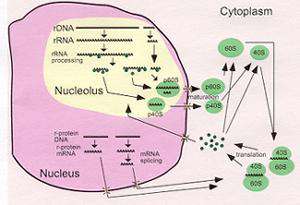
Ribosomes are the universal machines for translating the genetic code into protein. The basic organization and function of ribosomes is preserved in all organisms. Since the major macromolecular constituent of all cells is protein, cells require large numbers of ribosomes, especially during periods of rapid growth.
The major focus of our work are two aspects of ribosome biogenesis. First, we are studying how mature rRNA molecules are formed from the primary precursor transcript in Saccharomyces cerevisiae, a process which requires many nucleolar RNA and protein components. In particular we investigate the structure and function of RNase MRP, an RNA protein particle which cleaves the precursor rRNA. The RNA subunit of RNase MRP has many similarities with the RNA subunit of RNase P, an enzyme involved in processing of precursor tRNA. In addition the two RNA containing enzymes share eight of their nine protein subunits. We are pursuing comparative studies of these two RNA-protein enzymes to understand structure and function of the class of processing enzymes.
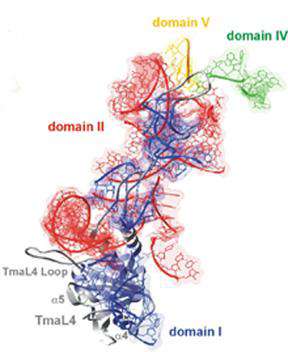
Second, we investigate transcription and translation regulation of ribosomal protein genes in bacteria. Encoded by the third gene of the eleven gene S10 operon in Escherichia coli, protein L4 is both a regulatory protein and a ribosomal protein. Recently, genetic studies in our lab and crystallography by other groups have pinpointed several sites in L4 for RNA protein interaction that are key determinants in regulation of the S10 operon, assembly of ribosomes and possibly ribosome function. Current work focuses on dissecting the role of the different domains of L4 and the evolution of the regulatory mechanisms for regulation of ribosomal protein synthesis.
Publications
Lindahl, L .(2022) Increasing Complexity of Ribosomes and Their Biogenesis. Int. J. Mol. Sci. 2022, 23, 8264. https://doi.org/10.3390/ijms23158264
Shamsuzzaman, M., Rahman, N., Gregory, B., Bommakanti, A., Zengel, J.M., Bruno, V. M. and Lindahl, L. Inhibition of Ribosome Assembly and Ribosome Translation Has Distinctly Different Effects on Abundance and Paralogue Composition of Ribosomal Protein mRNAs in Saccharomyces cerevisiae. mSystems 2023 https://doi.org/10.1128/msystems.01098-22
Li, X. Zengel, J.M., and Lindahl, L. A Novel Model for the RNase MRP-Induced Switch between the Formation of Different Forms of 5.8S rRNA. Int. J. Mol. Sci. 2021, 22, 6690. https://doi.org/10.3390/ijms22136690
Rahman N, Shamsuzzaman M, Lindahl L (2020) Interaction between the assembly of the ribosomal subunits: Disruption of 40S ribosomal assembly causes accumulation of extra-ribosomal 60S ribosomal protein uL18/L5. PLoS ONE 15(1): e0222479. https://doi.org/10.1371/journal.pone.0222479
Fox, J.M., R.L. Rashford, and Lasse Lindahl. 2019. Co-assembly of 40S and 60S ribosomal proteins in early steps of eukaryotic ribosome assembly. International Journal of Molecular Science. 20:2806; https://doi.org/10.3390/ijms20112806
Gregory B., Rahman N., Bommakanti A., Shamsuzzaman M., Thapa M., Lescure A., Zengel J.M., Lindahl L. 2019. The small and large ribosomal subunits depend on each other for stability and accumulation. Life Science Alliance. PMID: 30837296. DOI: 10.26508/lsa.201800150 [PDF]
Shamsuzzaman, M., A. Bommakanti, A. Zapinsky, N. Rahman, C. Pascual, and Lasse Lindahl. 2017. Analysis of cell cycle parameters during the transition from unhindered growth to ribosomal and translational stress conditions. PLoS ONE 12(10): e0186494.https://doi.org/10.1371/journal.pone.0186494
Wekselman, I., E. Zimmerman, C. Davidovich, M. Belousoff, D. Matzov, M. Krupkin, H. Rozenberg, A. Bashan, G. Friedlander, J. Kjeldgaard, H. Ingmer, L. Lindahl, J. M. Zengel, and A. Yonath. 2017. The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel. Structure 25:1-9. http://dx.doi.org/10.1016/j.str.2017.06.004; PMID:28689968
Nenad Ban,Roland Beckmann, Jamie HD Cate, Jonathan D Dinman, François Dragon, Steven R Ellis,6Denis LJ Lafontaine, Lasse Lindahl, Anders Liljas, Jeffrey M Lipton, Michael A McAlear, Peter B Moore, Harry F Noller, Joaquin Ortega,1Vikram Govind Panse, V Ramakrishnan, Christian MT Spahn, Thomas A Steitz, Marek Tchorzewski, David Tollervey, Alan J Warren, James R Williamson, Daniel Wilson, Ada Yonath, and Marat Yusupov. 2015. A new system for naming ribosomal proteins. Curr Opin Struct Biol.: 165–169.
[Abstract] [PDF]
Mamata Thapa, Ananth Bommakanti, Md Shamsuzzaman, Brian Gregory, Leigh Samsel, Janice M. Zengel, and Lasse Lindahl. 2013. Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes. Mol. Biol. Cell mbc.E13-02-0097; First Published on October 9, 2013; doi:10.1091/mbc.E13-02-0097. [Abstract] [PDF]
Lindahl L, A. Bommankanti, X. Li, L. Hayden, A. Jones, M. Khan,T. Oni, and J.M. Zengel 2009. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA, 15:1407-1416
[Abstract] [PDF]
Lawrence M, L. Lindahl, J.M. Zengel. 2008. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol. 190:5862-5869
[Abstract] [PDF]
Bomakanti, A.S., L. Lindahl, and J.M. Zengel. 2008. Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Saccaromyces cerevisiae. RNA, 14:460-464
[Abstract]
Zaman, S.M., L. Lindahl, and J.M. Zengel. 2007. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in E. coli. Mol. Microbiology, 66:1039-1050.
[Abstract]
Allen T.D., T. Watkins, L. Lindahl, and J.M. Zengel. 2004. Regulation of ribosomal protein synthesis in Vibrio cholerae. J Bacteriol. 186:5933-7.
[Abstract]
Li X, S. Zaman, Y. Langdon, J.M. Zengel and L. Lindahl. 2004. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 32:3703-11.
[Abstract]
Zengel, J.M. and L. Lindahl. 2003. Assay of transcription termination by ribosomal protein L4. Methods Enzymol. 371:356-69.
[Abstract]
Zengel, J.M., A. Jerauld, A.Walker, M.C. Wahl and L. Lindahl. 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA 9:1188-1197.
[Abstract]
Stelzl, U., J.M. Zengel, M. Tovbina, M. Walker, K.H. Nierhaus, L. Lindahl, and D.J. Patel. 2003. RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J. Biol. Chem. 278:28237-28245.
[Abstract]
