Senior Research Scientist

| Office: | BS 457 |
|---|---|
| Phone: | 410-455-2976 |
| Lab: | BS 251 |
| Lab Phone: | 410-455-3120 410-455-3572 |
| Email: | zengel@umbc.edu |
Education
Postdoctoral, Baylor College of Medicine, 1978
Postdoctoral, Stanford University, 1977
Ph.D., University of Wisconsin, Madison, 1976
B.A., McDaniel College, 1970
Professional Interests
Research in my laboratory focuses on the formation and function of ribosomes, the cellular machines responsible for protein synthesis. We are investigating various aspects of ribosome synthesis, structure, and function, using genetic, biochemical, and phylogenetic approaches to investigate these processes at the molecular level.
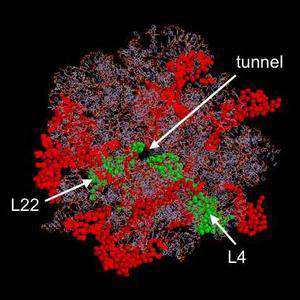
One focus in the lab is on the role of ribosomal proteins. Although several key functions of ribosomes are catalyzed by the rRNA, there is persuasive evidence that ribosomal proteins play a key role in the assembly and function of ribosomes. Our laboratory has developed methods in the eubacteria Escherichia coli and Deinococcus radiodurans and in the yeast Saccharomyces cerevisiae for in vivo synthesis of 50S ribosomal subunits containing mutant ribosomal protein L4 or L22. Our work is focused particularly on versions of L4 or L22 with deletions, insertions, or point mutations in the tentacle-like region of the proteins that extends from the globular surface domain to the peptide exit tunnel. The tips of these tentacles, which form part of the lining of the peptide exit tunnel, have been implicated in a gating mechanism that regulates the rate of translation. To complement our site-directed mutagenesis studies, we have isolated mutations in the L4 and L22 genes of both E. coli and D. radiodurans that bestow resistance to macrolide antibiotics such as erythromycin and troleandomycin. We are now characterizing the effects of these mutations on cell growth and on ribosome assembly, function, and structure. We are also analyzing similar tentacle mutations in the corresponding ribosomal proteins of S. cerevisiae.
Another focus in the lab is the processing of rRNA in the yeast S. cerevisiae. We have shown that RNase MRP, an endonuclease found in all eukaryotes, is involved in multiple steps in processing the primary rRNA precursor transcript. This enzyme contains one RNA and eight-to-ten protein subunits, and has many similarities with the tRNA-processing enzyme RNase P. To better understand the function of RNase MRP, we are isolating and characterizing mutations in the various components of the enzyme. We are also developing methods for purifying the enyzyme so that we can analyze its activity in vitro.
Publications
Lawrence M, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol. 2008. [Epub ahead of print]
[Abstract] [PDF]