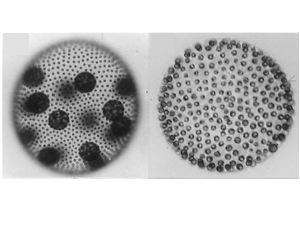
Our group uses the multicellular green alga Volvox carteri to study mechanisms of cellular differentiation, and how they evolve. V. carteri possesses ~2000 cells but just two cell types: large reproductive cells called gonidia, and small, motile somatic cells. Gonidial and somatic cell precursors are set aside when a subset of blastomeres undergo visibly asymmetric cell divisions during embryogenesis. The larger progeny produced by these divisions become reproductive cells, or gonidia, which act as asexual stem cells that ultimately produce the next generation of gonidia and somatic cells, while the smaller cells ultimately differentiate into bi-flagellate somatic cells that provide the organism motility but never divide.
Most research in our lab aims to better understand two important aspects of V. carteri development. First, we want to know how asymmetric cell divisions are regulated—why certain cells undergo them while others do not, and how the division plane is shifted off center to produce cells that differ in size. Proper control of cell division symmetry is crucial for all plants and animals to develop normally, yet relatively little is known about the mechanisms that determine where division planes are placed. We are using V. carteri mutants defective for asymmetric division (called Gonidialess, because they fail to make large cells that can become gonidia) to clone genes required for asymmetric division; analysis of these genes should yield important insights into how asymmetric division is controlled in V. carteri. So far we have cloned one gls gene, glsA, which encodes an embryo-specific, J-protein chaperone (GlsA) that localizes to the nucleus and is ~equally abundant in all blastomeres, those that divide symmetrically as well as asymmetrically. GlsA interacts with another chaperone, Hsp70A, which is enriched in the blastomeres that cleave asymmetrically. We hypothesize that GlsA and Hsp70A regulate the activities of transcription factors that control downstream genes required for asymmetric division, and we also hypothesize that relative Hsp70A abundance determines which cells will divide asymmetrically. Current projects in the lab aim to reveal how Hsp70A becomes asymmetrically distributed in the embryo, to identify and characterize other proteins that GlsA interacts and functions with, to clone and characterize additional gls genes.
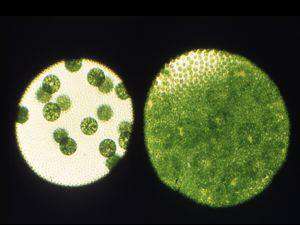
Our second major goal is to understand how the somatic cell fate is maintained in V. carteri, and how it evolved. The key to these studies is the somatic regenerator, or regA gene. regA mutants develop normally through the first day of the two-day life cycle, forming gonidia and somatic cells as in the wild type, but the somatic cells dedifferentiate, enlarge, and become gonidia. Thus regA is required to maintain the repression of growth and cell division in somatic cells. regA encodes a nuclear protein (RegA) that is enriched in the types of amino acids normally abundant in transcriptional repressors, and it also possess a ~110-aa VARL domain (Volvocine Algal RegA like) that resembles the DNA-binding SAND domain that has been identified in a number of plant and animal transcription factors. We are testing the idea that RegA can repress transcription, and we also aim to determine if it can bind DNA, and if so, what its direct targets are. In addition, we are analyzing several regA-related genes from V. carteri and the related unicellular alga, Chlamydomonas reinhardtii. Learning more about these genes, and about regA, should tell us how certain cells differentiate as, and remain as somatic cells, and how the somatic cell fate arose in the first place.
Projects
Identification and characterization of binding partners of the asymmetric division protein GlsA
We have screened a yeast-two hybrid library of V. carteri polypeptides for candidate interacting partners of GlsA. One of the candidates, Hsp70A, has since been shown to be the only cytoplasmic Hsp70 in V. carteri, and to bind GlsA as a chaperone partner in Volvox and to be required for asymmetric division. Current efforts are focused on analyzing other candidates from the screen, in particular to determine which if any are required for asymmetric cell division.
Current Members
| Robin Bridgman Graduate Student: Ph.D., Biological Sciences |
 |
| J.D. Seah Graduate Student: Ph.D., Biological Sciences |
 |
| James Williams Graduate Student: Ph.D., Biological Sciences |
 |
Former Members
|
Manar Adbelkader
Undergraduate Researcher |
 |
| Audrey Allison Lab Support Student |
 |
|
Eric Balzer
B.S., Biological Sciences, UMBC Eric is currently a second-year Ph.D. student in the department of Molecular and Cell Biology at the University of Maryland, Baltimore, in the laboratoryof Dr. Stuart Martin, where he is studying the role of cytoskeltal factors in breast cancer metastasis. |
 |
|
Christopher Bednarek
Undergraduate Researcher
The green alga Volvox carteri possesses just two cell types and is an excellent model for the study of cellular differentiation mechanisms and their evolution. Currently there are only two selectable markers available for introducing genes into V. carteri to study their function. To create a new selectable marker for V. carteri transformation, a chlorate-resistant nitrate reductase gene (CR-nitA), has been synthesized. Nitrate reductase reduces nitrate to nitrite and, similarly, chlorate to chlorite, which is toxic to cells. Previous studies have demonstrated that a cysteine to alanine change in a conserved amino acid in nitrate reductase yields an enzyme that dimerizes but does not retain enzymatic activity. The CP-nitA marker gene encodes a product with an analogous c139a mutation and is driven by a high expression promoter so that transformants that take it up should over-express non-functional CP-NitA protein that dimerizes with wild type nitrate reductase. This should titrate functional nitrate reductase protein and permit transformants to survive chlorate. I am using gene assembly and subcloning to synthesize a Volvox CP-nitA gene with a constitutive Hsp70-Rubisco-gene promoter and an HA-epitope tag. I will test the completed construct for chlorate resistance following transformation into V. carteri.
|
 |
| Stephanie Buckley B.S., Biological Sciences, UMBC Stephanie is the recipient of a 2006 NIH Post-Baccalaurate Intramural Research Training Award (IRTA) and is currently a trainee in this program. |
 |
| Qian Cheng Graduate Student: Ph.D. Qian received her Ph.D. in August, 2005 and is now a postdoctoral fellow in the laboratory of Dr. Jiandong Cheng at the H. Lee Moffitt Cancer Center & Research Institute in Tampa, Florida. Currently she is investigating the E3 ligase that targets the tumor suppressor protein p53. |
 |
| Kori Conyers B.S., Biological Sciences, UMBC Kori is currently a first-year student in the Medical Scientist Training Program at UT Southwestern Medical Center in Dallas, Texas. |
 |
| Tylen Darling Undergraduate Researcher |
 |
|
Wipawee Dejtisakdi
|
 |
| Anna Gitterman Undergraduate Researcher |
 |
| Seung-Ho Lee Undergraduate Lab Assistant |
 |
| Stephanie Owusu Undergraduate Researcher |